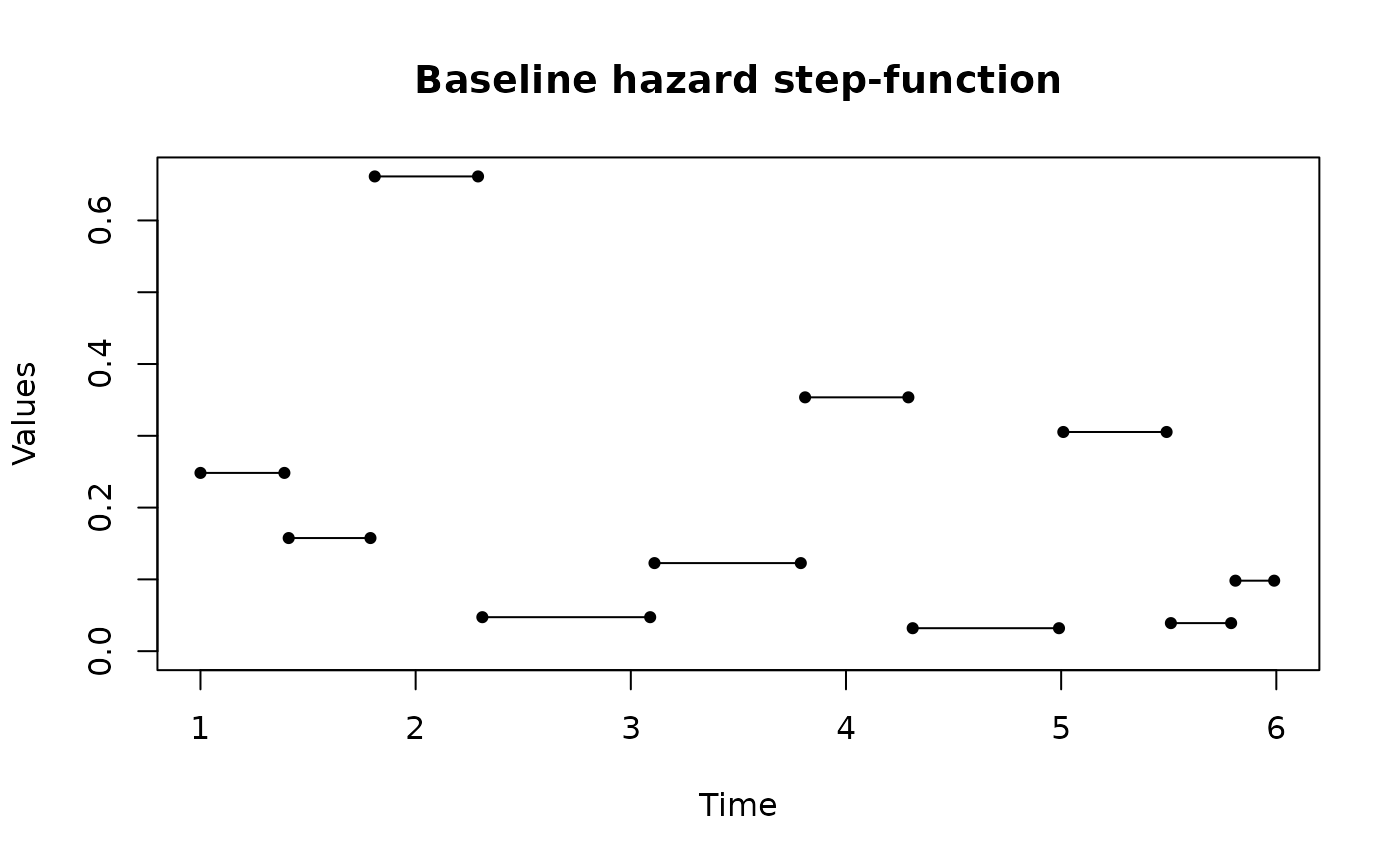
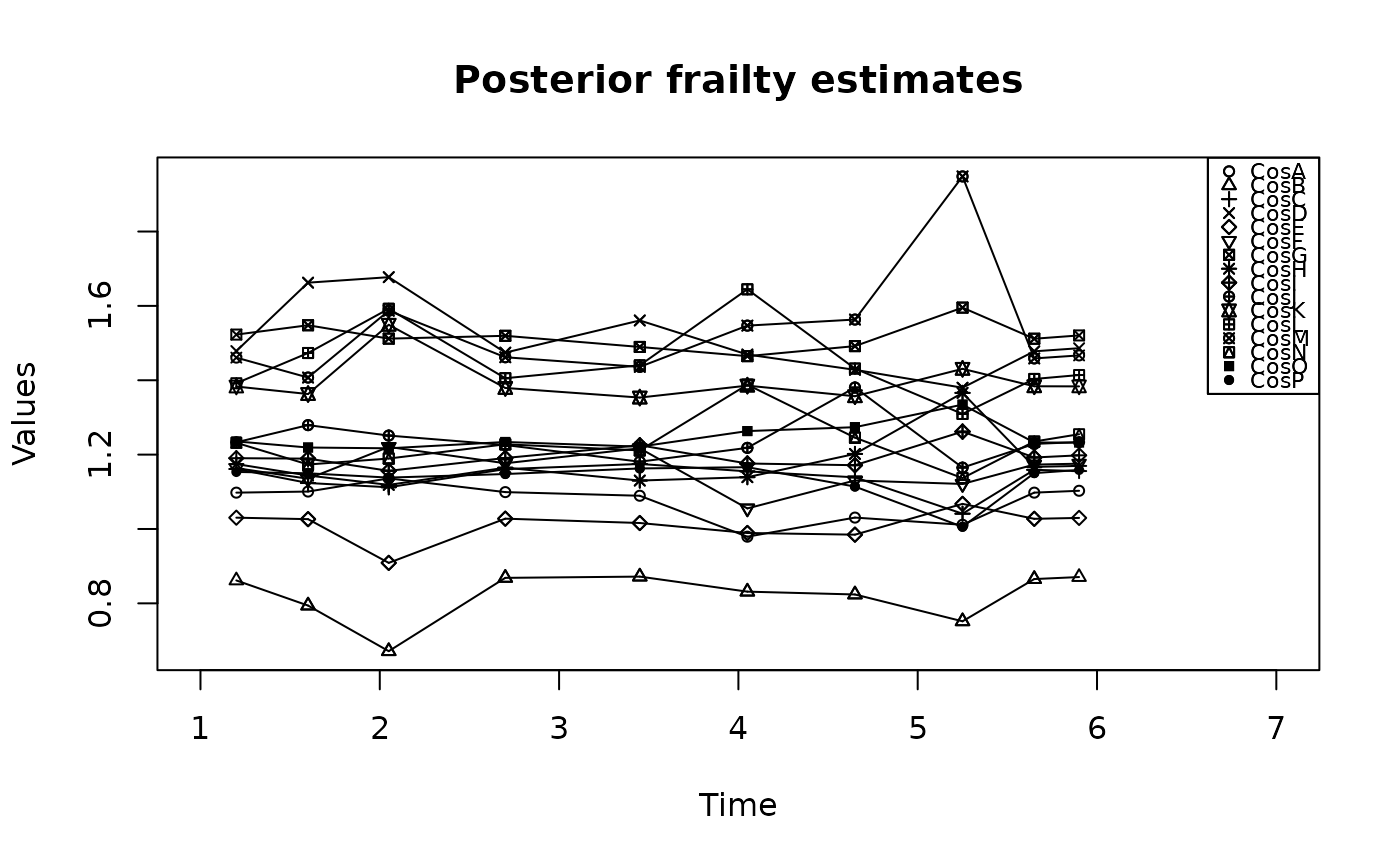
Plots Related to the the 'Adapted Paik et Al.' Model
plot.AdPaik.RdPlots Related to the the 'Adapted Paik et Al.' Model
Examples
# Import data
data(data_dropout)
# Define the variables needed for the model execution
eps_paik <- 1e-10
categories_range_min <- c(-8, -2, eps_paik, eps_paik, eps_paik)
categories_range_max <- c(-eps_paik, 0.4, 1 - eps_paik, 1, 10)
time_axis <- c(1.0, 1.4, 1.8, 2.3, 3.1, 3.8, 4.3, 5.0, 5.5, 5.8, 6.0)
formula <- time_to_event ~ Gender + CFUP + cluster(group)
# Call the main model function
# \donttest{
result <- AdPaikModel(formula, data_dropout, time_axis, categories_range_min, categories_range_max)
plot(result)
 # }
# }